Metagenomics is the study of all microbial genomes present in an environment, from the bottom of the sea to inside our own bodies. If you have shotgun NGS sequencing data, Partek provides powerful, easy-to-use analysis tools to characterize microbial communities.
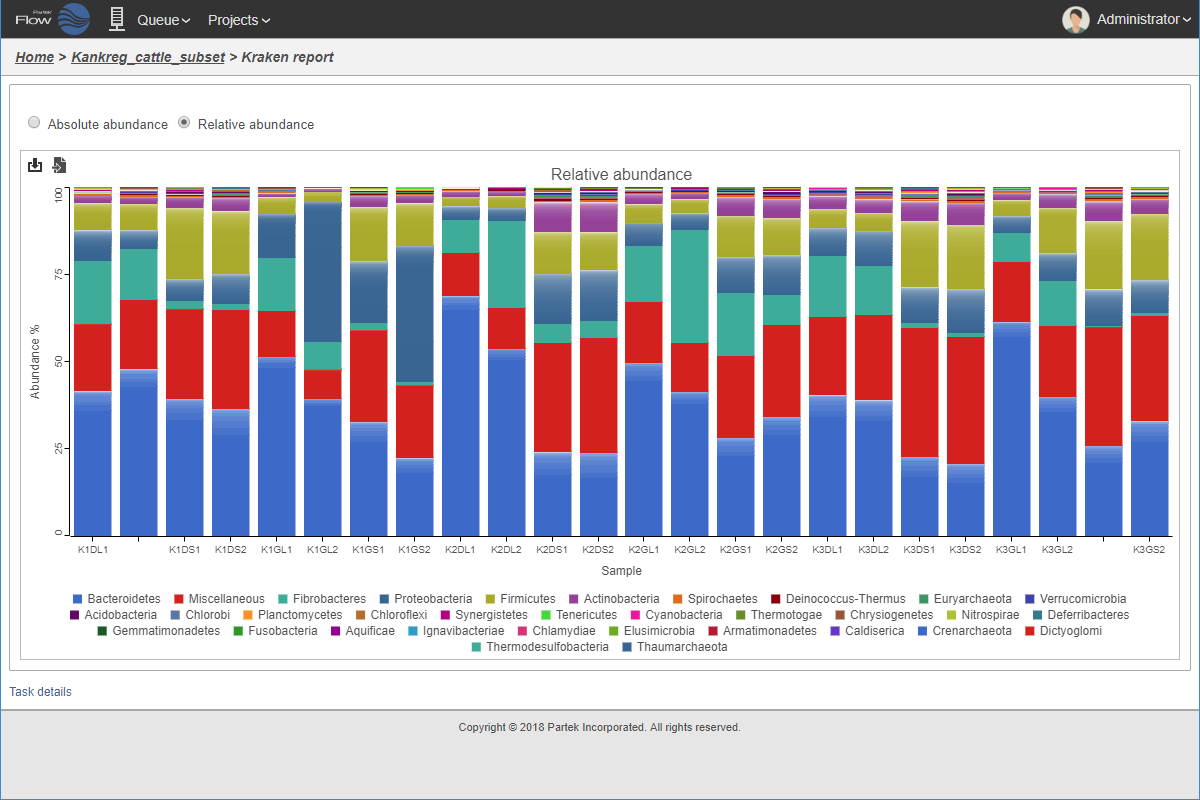
Relative taxonomic abundance plot.
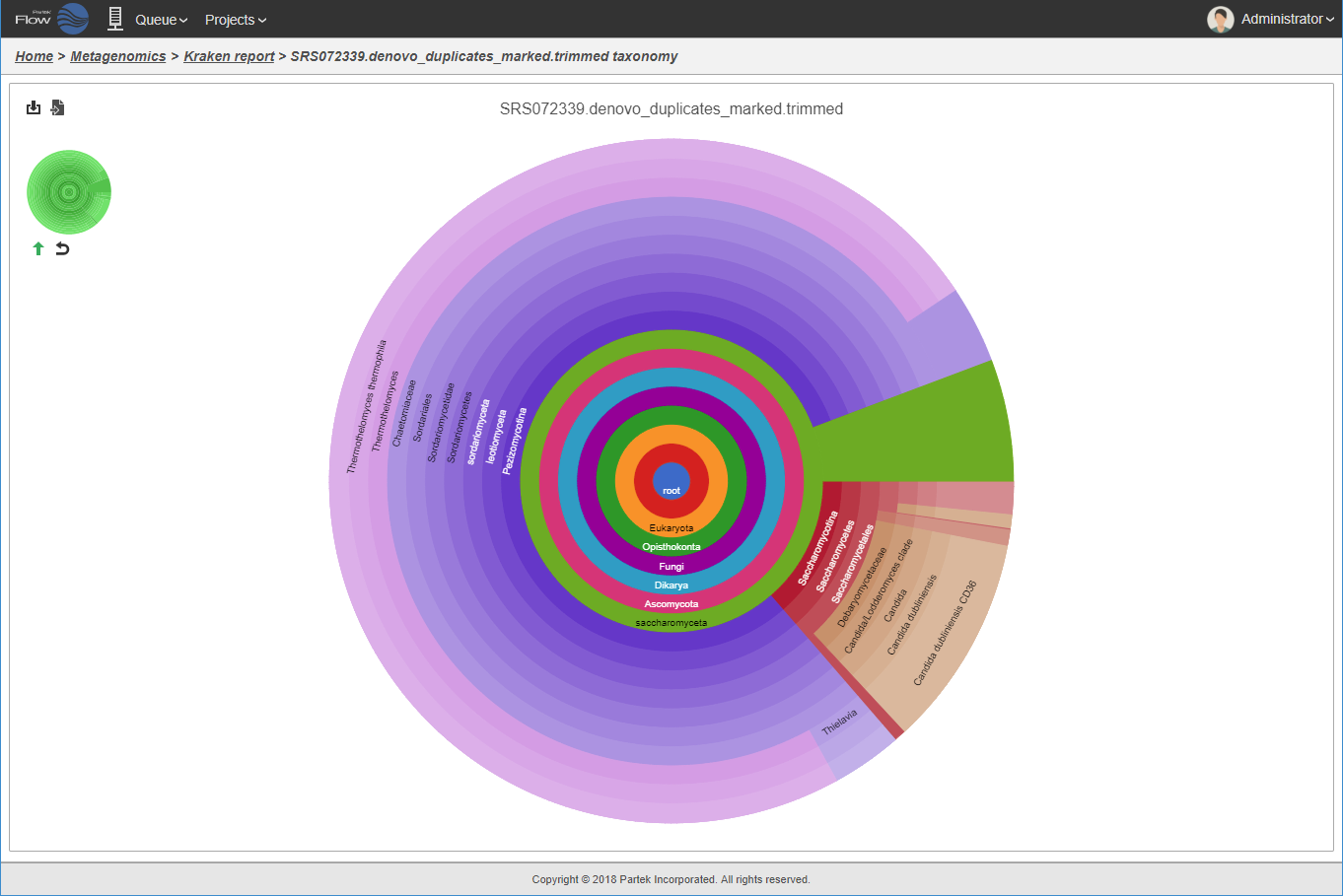
Interactive hierarchical pie chart.
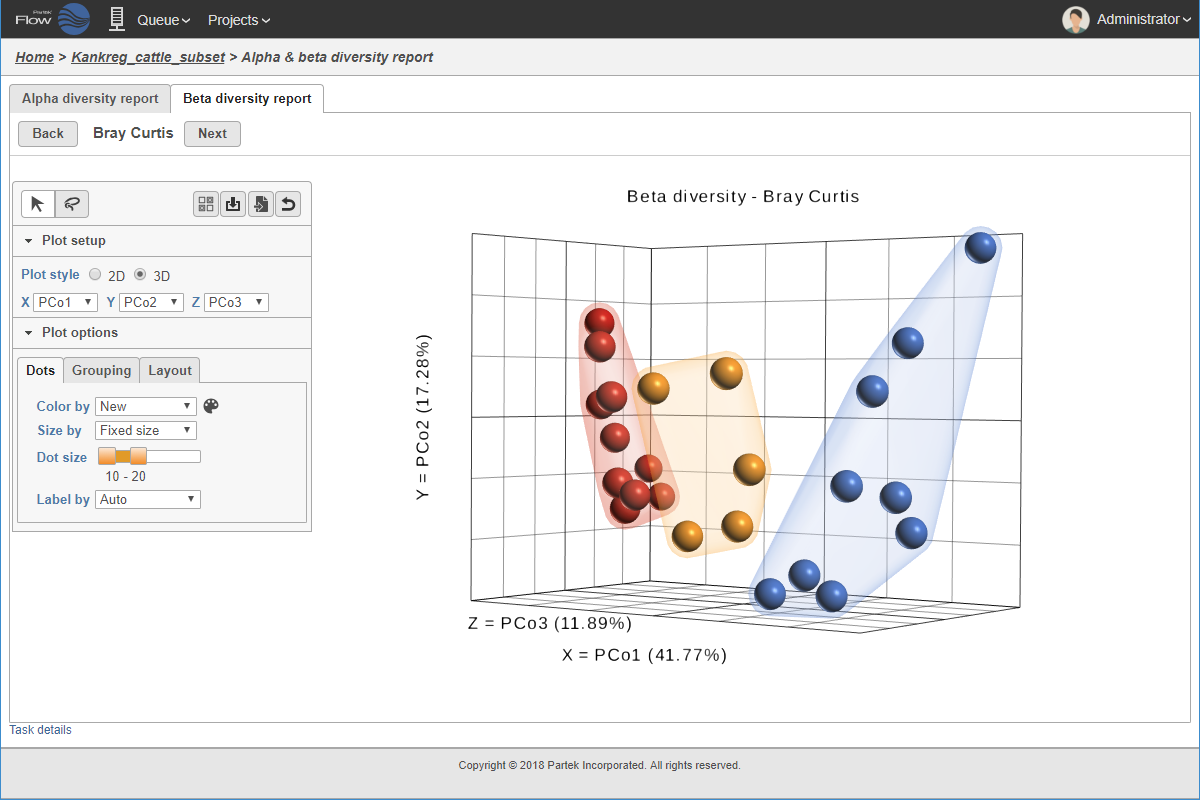
PCoA analysis.

QA/QC

Host DNA Removal


Taxonomic Classification


Alpha Beta Diversity

Fast and Accurate Identification of Taxonomic Origins
Metagenomic data sets consist of millions of short NGS reads, which originate from different microbial genomes. Sorting through such a large, fragmented data set to find which organism each read originates is not only complex, but computationally challenging. Partek Flow makes use of the ultrafast metagenomic sequence classification tool Kraken. This tool leverages public databases containing thousands of microbial reference sequences and accurately identifies the taxonomic origin of each sequencing read.
Advanced Statistics Identify Microbial Diversity
One fundamental question in metagenomics analysis is: how does microbial diversity differ between groups? One approach is to measure alpha (within-sample) diversity and use this to describe diversity changes. However, this approach lacks statistical rigor and does not account for different study designs. In Partek, you can use our powerful, flexible ANOVA statistics to look for meaningful changes in microbial diversity to better understand the patterns of microbial diversity in your study.

Customizable and Flexible Sequence Search
Everyone has a different research question. You may be trying to learn what type of Respirovirus is present in mixed clinical samples, or how bacterial communities change in response to different diets. Thus, there is a need to tailor the data analysis to the research question. With the flexible library file management tools in Partek Flow, you can create a custom database to perform your metagenomic sequence search against. This allows you to fine-tune your search or make it as broad as possible.
Selected Metagenomic Publications Citing Partek Software
Functional genome and microbiome in blood of goats affected by the gastrointestinal pathogen Haemonchus contortus
Tilahun, Yonathan et al., bioRxiv (2021)
Age associated microbiome modulation and its association with systemic inflammation in a Rhesus Macaque Model
Pallikkuth, Suresh et al., bioRxiv (2020)

Ask a question or request a software demonstration/trial
