Our understanding of the functional importance of non-coding RNA (ncRNA) is growing. Several classes exist, including microRNA (miRNA), long non-coding RNA (lncRNA), piwi-interacting RNA (piRNA) and others. Whether you have NGS or microarray data, Partek provides easy-to-use software that guides you through the analysis from start to finish.
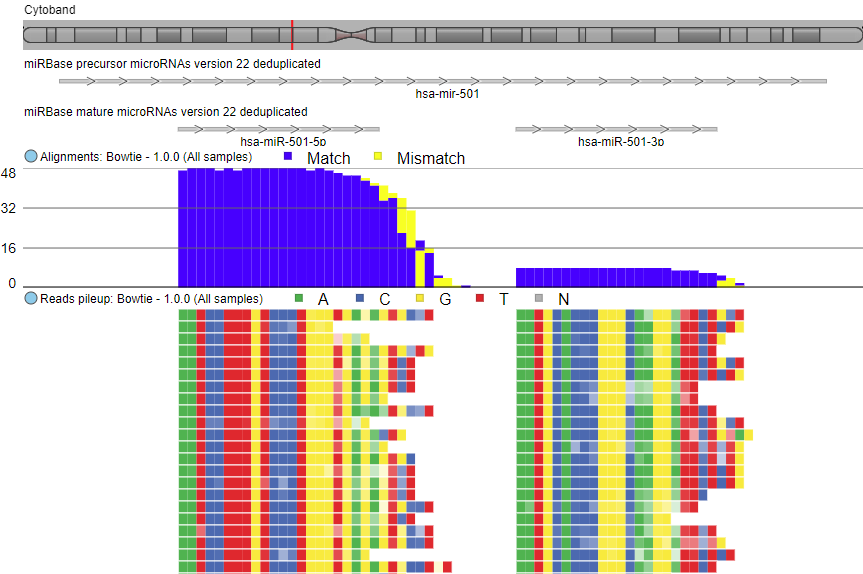
Align NGS reads with optimal settings.
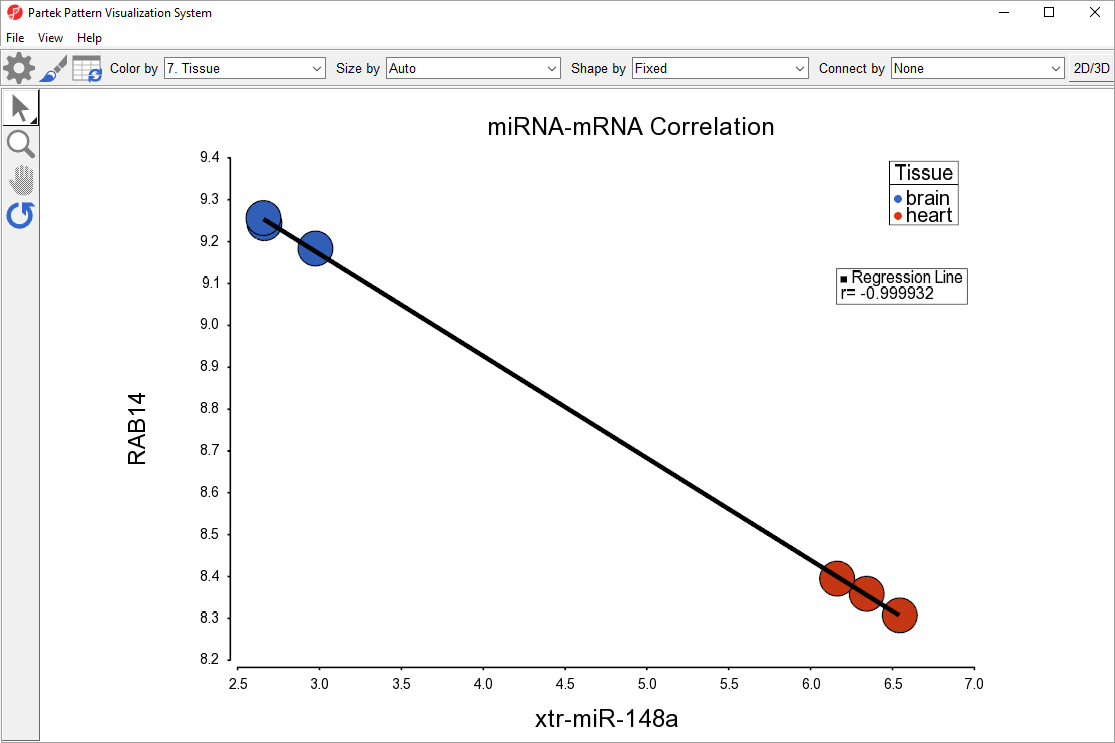
Correlate miRNA and mRNA expression for novel discovery.
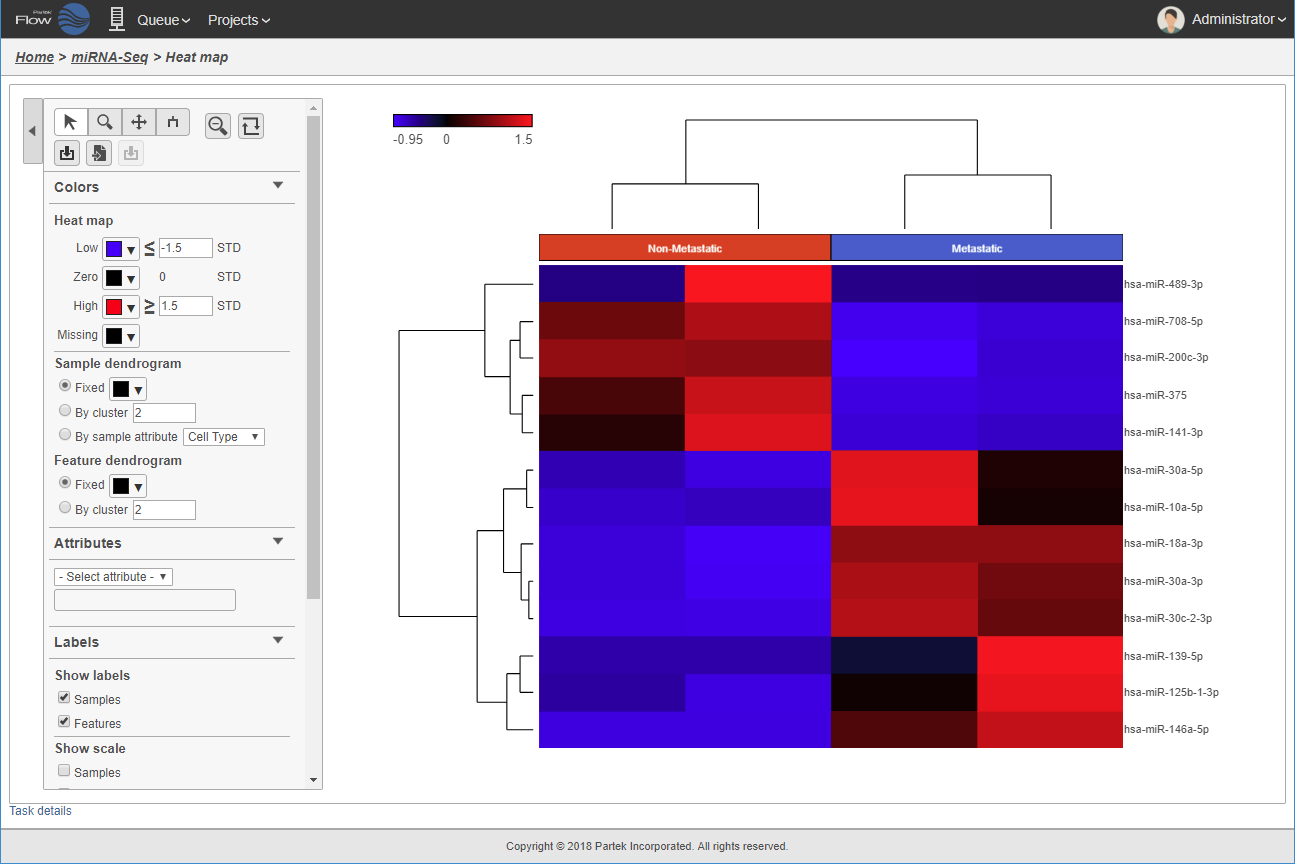
Differential analysis for biomarker discovery.

Alignment


QA/QC


Quantification

Differential Analysis

Data Integration

Flexible Library Management for Up-to-Date Databases
As our understanding of ncRNA grows, so do publicly available databases. Keeping up to date with the latest knowledge base is challenging and is further complicated by different databases storing information in different formats. Partek software makes the process easy with a flexible library file management system, that lets you automatically download the latest annotation databases or add your own custom ncRNA from any source, in any format (GTF, GFF3, BED and more).

Mine Public Databases and Integrate RNA Datasets
Partek software provides easy to use analytical solutions to mine public databases to find potential mRNA targets for lists of ncRNAs. Furthermore, ncRNA and gene expression data can be integrated to facilitate novel functional discovery. ncRNAs function to regulate gene expression across the transcriptome. Our software helps you understand the interactions between ncRNA and mRNA expression.

Powerful and Reliable Statistics for Biomarker Discovery
Because of the ubiquity and fundamental importance of ncRNA in biological systems, there is increasing interest in using these molecules as biomarkers for disease. An important first step is to find ncRNAs that are differentially expressed in different groups of samples. This requires powerful and reliable statistics, such as those found within Partek software, to precisely identify the best candidates. Partek software uses a combination of custom and public domain statistical algorithms which are easy to use for statisticians and non-statisticians alike.

Selected non-Coding RNA Expression Publications Citing Partek Software
A blood miRNA signature associates with sporadic Creutzfeldt-Jakob disease diagnosis
Norsworthy, Penny J. et al., Nature Communications (2020)
Variability in cerebrospinal fluid MicroRNAs through microRNAs
Prieto-Fernández, Endika et al., Molecular Neurobiology (2020)
Small RNA profiling of piRNAs in colorectal cancer identifies consistent overexpression of piR-24000 that correlates clinically with an aggressive disease phenotype
Narayanan Iyer, Deepak et al., Cancers (2020)
Early microRNA indicators of PPARα pathway activation in the liver
Chorley, Brian N. et al., Toxicology Reports (2020)
In silico analysis of RNA and small RNA sequencing data from human BM-MSC’s and differentiated osteocytes, chondrocytes and tenocytes
Koduru, Srinivas V. et al., Engineered Regeneration (2021)

Ask a question or request a software demonstration/trial
