Cytosine methylation is a process by which a methyl group is added to cytosine, typically at CpG sites. It is a key epigenetic mechanism involved in the regulation of gene expression and drives numerous biological processes such as development, aging, carcinogenesis, and genomic imprinting. Starting with raw data from the Infinium™ MethylationEPIC or Infinium™ Human Methylation 450K BeadChip array, you can use Partek tools to perform every analysis step from differential methylation to biological interpretation.
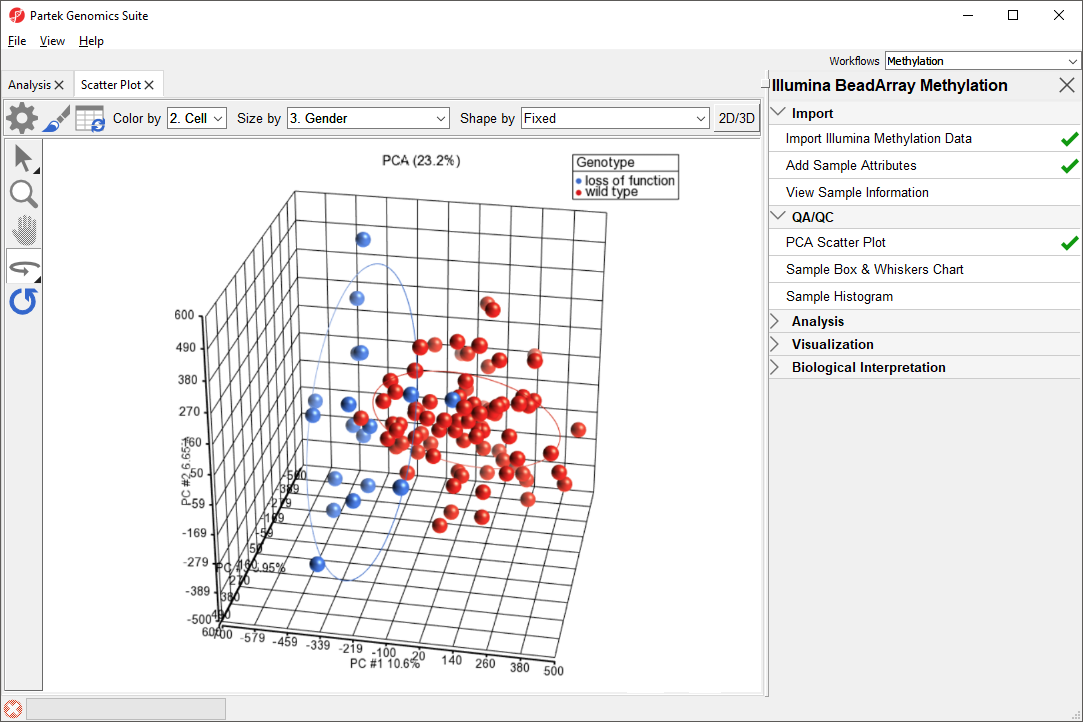
Methylation patterns between groups.
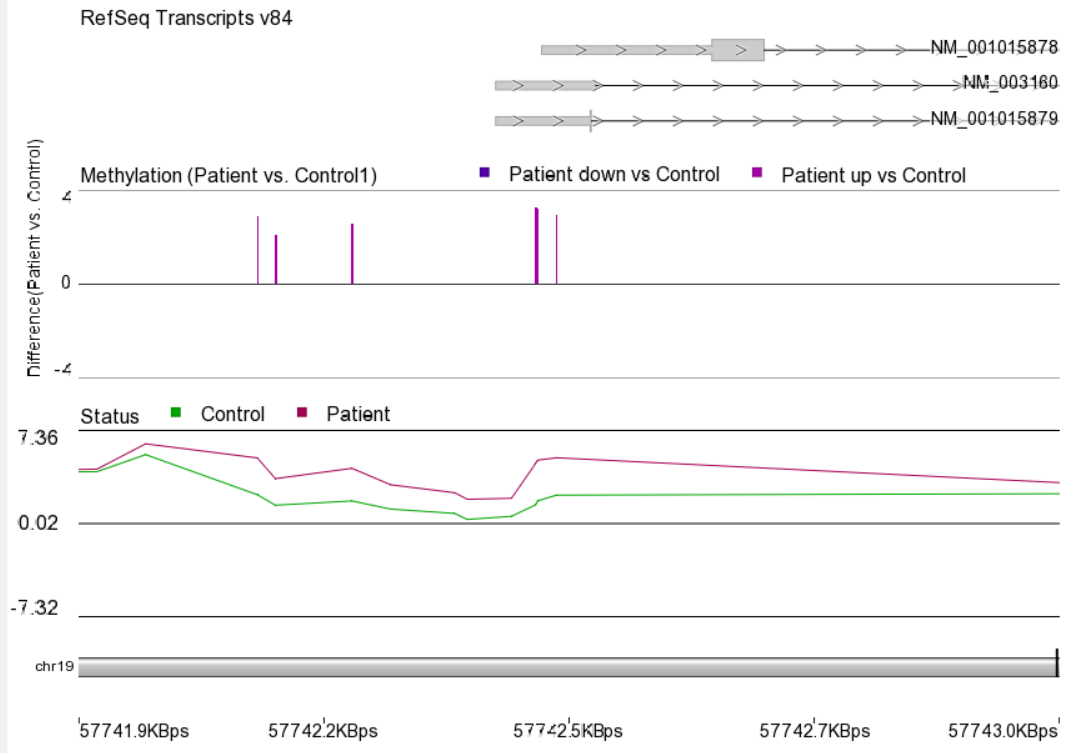
Differentially methylated CpG sites.
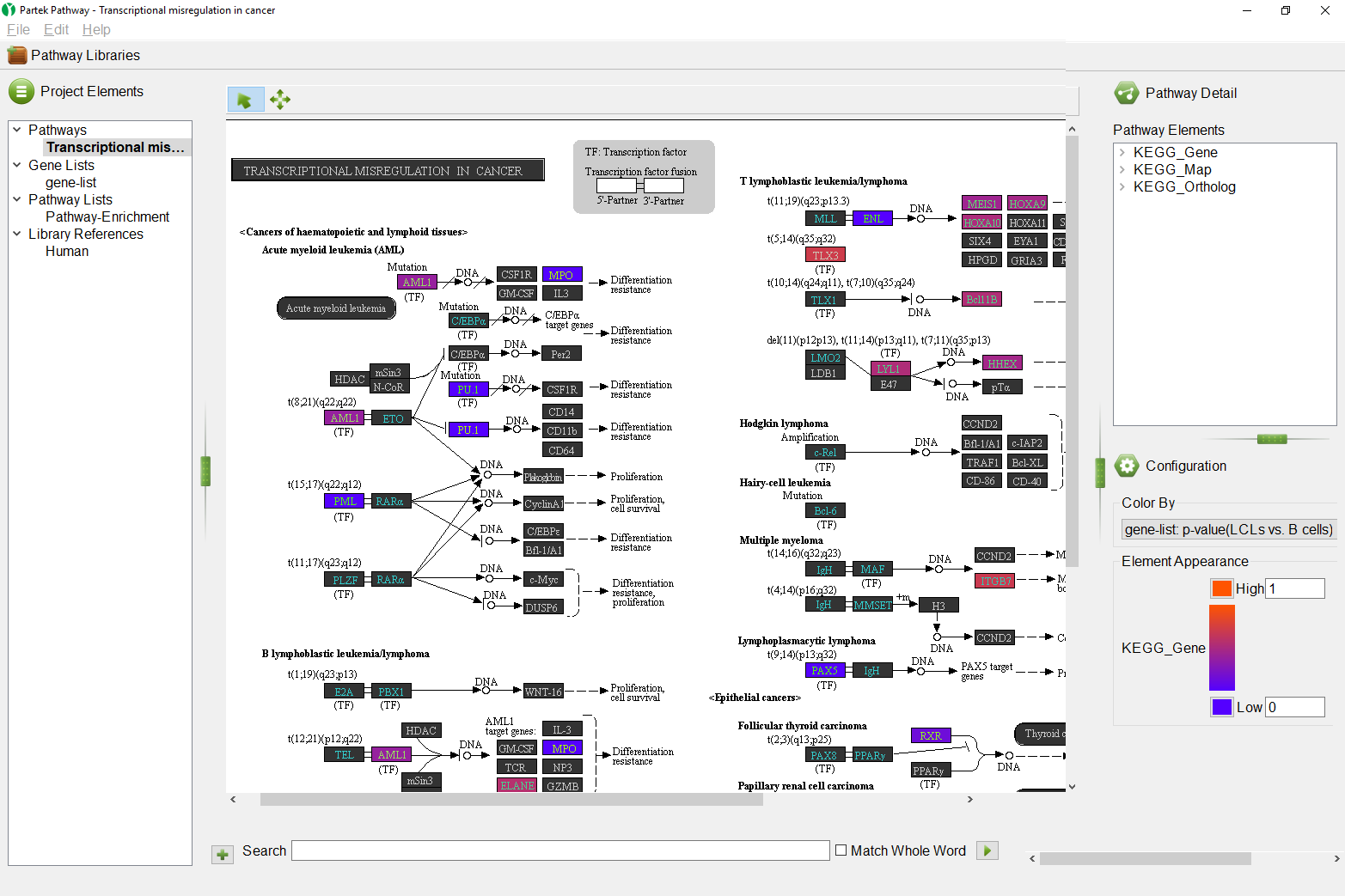
Pathways regulated by methylation.
Normalization


QA/QC

Differential Analysis

Biological Interpretation

Methylation QA/QC
You cannot expect to obtain valid results if your data is not technically sound. Since there are many metrics that could potentially be considered, performing QC of methylation data is not always straightforward. Partek tools allow you to choose between key and easy-to-interpret QC output. You can also interrogate your data at a granular level for an in-depth report.
Integrate Methylation Data with Gene Expression
A common motivation to study methylation analysis is to learn about the association between methylation levels and gene expression. Using Partek tools you can perform analysis of each of those data sets independently, as well as integrate the results and detect genes that show evidence of both differential methylation and differential expression.

Selected Methylation Publications Citing Partek Software
High glucose alters the DNA methylation pattern of neurodevelopment associated genes in human progenitor cells in vitro
Kandilya, Deepika et al., Scientific Reports (2020)
The comparative methylome and transcriptome after change of direction compared to straight line running exercise in human skeletal muscle
Maasar, Mohd-Firdaus et al., Frontiers in Physiology (2021)
Effect of early parenteral nutrition during paediatric critical illness on DNA methylation as a potential mediator of impaired neurocognitive development: a pre-planned secondary analysis of the PEPaNIC international randomised controlled trial
Güiza, F. et al., The Lancet (2020)
Assessment of differentially methylated loci in individuals with end-stage kidney disease attributed to diabetic kidney disease: an exploratory study
Smyth, L.J., Clinical Epigenetics (2021)
Transcriptome and methylome analysis reveals three cellular origins of pituitary tumors
Taniguchi-Ponciano, Keiko et al., Scientific Reports (2020)
DNA methylation across the genome in aged human skeletal muscle tissue and muscle-derived cells: the role of HOX genes and physical activity
Turner, D.C. et al., Scientific Reports (2020)

Ask a question or request a software demonstration/trial
