 Analyzing spatial transcriptomic data in Partek Flow.
Analyzing spatial transcriptomic data in Partek Flow.
We may all be under the impression that single cell RNA-Seq is a new technology, but it has been around for more than a decade (Tang F et al. Nat Methods 2009). Hard to believe, right? And what about tissue transcriptomics (a.k.a. spatially resolved transcriptomics)? If we focus only on RNA sequencing-based analysis, then the first paper was published in 2013 (Ke et al. Nat Methods 2013), which in the “omics” age, cannot be considered a novelty. To be fair, the concept of gene expression analysis within tissue context is considerably older—first introduced in 1982 (Singer & Ward. Proc Natl Acad Sci USA 1982). Either way, tissue transcriptomics really rose to prominence within the last two years and continues to gain a lot of attention.
Transcriptome-wide studies of gene expression certainly provide invaluable insight into biology on a molecular level, particularly when performed at the single cell level (i.e., single cell RNA-Seq). However, some key pieces of information such as the tissue relationships, are still missing and are only made available by tissue transcriptomics techniques. Knowing the location of each cell, as well as its surroundings, enable us to fully understand and appreciate molecular events. Two straightforward examples that come to mind are embryonic development and tumor microenvironments. One of the key features of the development is spatially and temporally coordinated gradients of gene expression, which then turn off developmental genes. Cancer tissue, the second example, is composed of different neoplastic populations and has a “contaminating” component of healthy tissue such as stroma or especially exciting, invading leukocytes.
Moreover, some tissues are not easily singularized (e.g., cartilage, bone, nervous tissue) or if they are, single cell suspension still lacks topographical information (e.g., bone marrow, lymph nodes). For instance, if you study interactions between bone and bone marrow the positional information is critical.
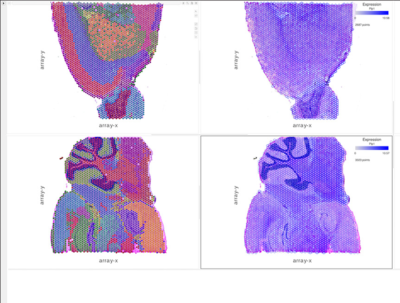
And finally, spatial considerations do not need to be limited to the tissue level but can also be transferred to the level of an individual cell, as intra-cellular functions and signals require well-defined positional arguments (Asp et al. in BioAssays 2020). This is exactly where tissue transcriptomics comes into play, as it merges gene expression data and spatial data. At the same time, we know what genes are expressed, their expression levels, and where within the tissue those genes are located.
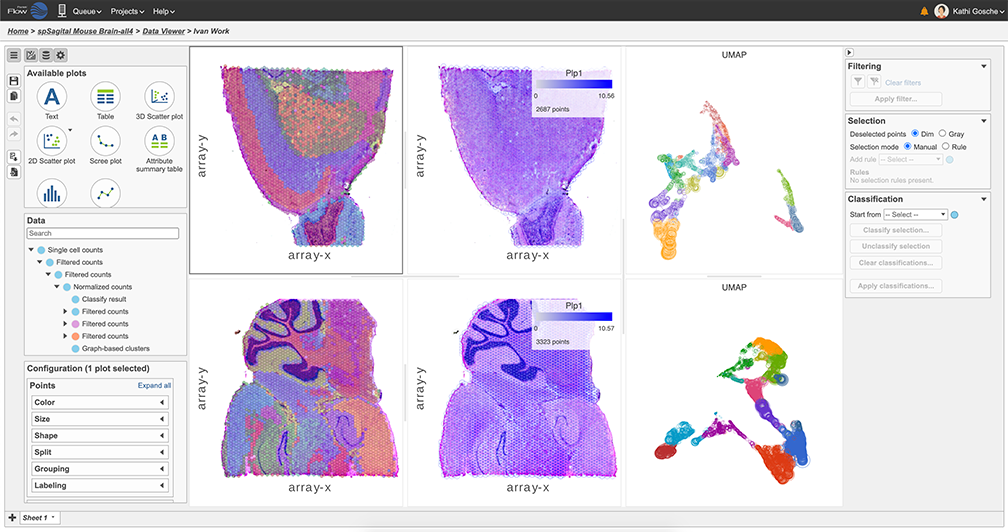
To learn how to perform tissue transcriptomics analysis in Partek Flow, take a look at our recent webinar: Single Cell Analysis – Tissue Transcriptomics. The webinar is based on 10X Genomics’ Visium data, but you are not limited to that assay: other related technologies, such as Nanostring’s GeoMx Digital Spatial Profiling are also supported.