Integration of Copy Number and Methylation Data from the Illumina® Infinium HumanMethylation450 BeadChip Array
Integration of genomic data and epigenomic data as a means for understanding complex genomic regulatory mechanisms is gaining in popularity. In this webinar, we will show you how to use Partek Genomics Suite to identify copy number regions from the Illumina® Infinium HumanMethylation450 BeadChip array and merge the results with differentially methylated regions detected using the same arrays.
Learn how to:
- Import Illumina .idat files and normalize the data
- Detect differentially methylated regions
- Remove probes with SNP in the vicinity of the query site
- Visualize the methylation signature
- Create a list of genes regulated by differential methylation
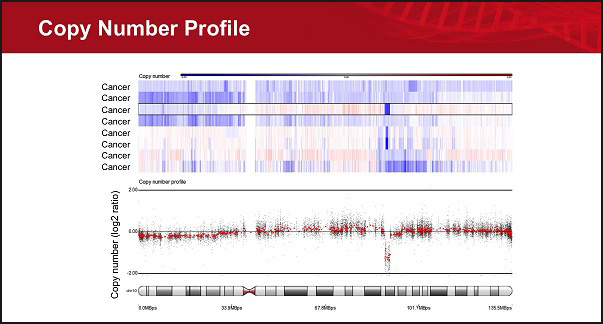
- Identify copy number regions
- Find copy number regions shared across study samples
- Integrate the copy number data with methylation data
Register to Watch
Share this post:
