Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology
A Research Spotlight Presentation
Research performed and presented by:
Victor Martinez, Ph.D.
Biomedical Sciences, British Columbia Cancer Research Centre
PIWI-interacting RNAs (piRNAs) are small (24-32bp) non-coding RNAs with a key role in epigenetic regulation of gene expression and maintenance of genomic stability in germ cells. Recent evidence suggests they are also expressed and functionally active in somatic tissues. The expression of individual piRNAs has been explored in some cancer types; however, their somatic expression patterns are largely uncharted.
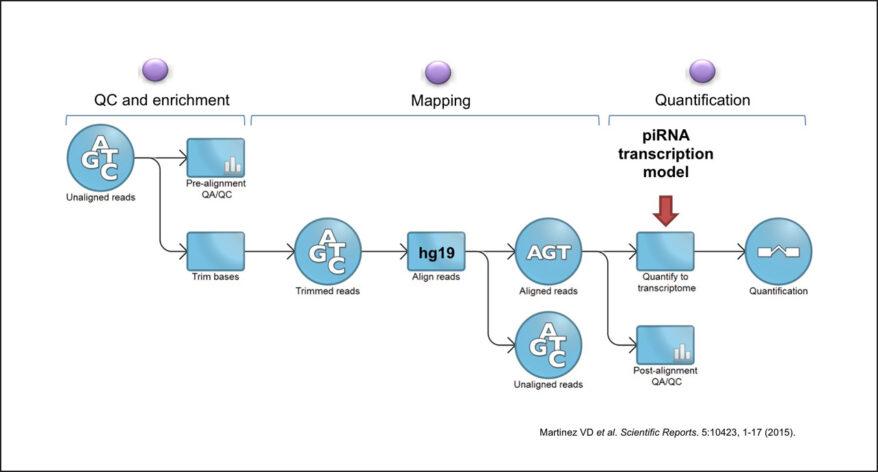
To investigate piRNA expression in human tissues, we analyzed 6,260 small non-coding RNAs (sncRNAs) sequence samples from 11 different organs available from The Cancer Genome Atlas. To deduce the expression of approximately ~32,000 human piRNAs on each sample, we built a custom small-RNA sequence analysis pipeline supported by the Partek Flow platform. This strategy allowed us to analyze one of the biggest cohorts of sncRNAs available worldwide, in a time- and cost-effective manner.
We discovered that a small, but significant fraction of the known piRNAs are expressed in somatic tissues, displaying tissue-specific patterns. Moreover, more than 500 piRNAs are expressed in corresponding tumour tissues, largely distinguishing tumour from non-malignant tissues in a cancer-type specific manner. Most expressed piRNAs mapped to known transcripts, contrary to “piRNA clusters” reported in germline cells. We also showed that piRNA expression signatures can delineate clinical features, such as histological subgroups, disease stages, and patient survival.
Here, we provide evidence of somatic, tissue-specific human piRNA expression. Aberrant expression patterns contribute to cancer subtype-specific biology. We discover piRNA-based signatures that identify aggressive tumors and have prognostic value. The unique expression patterns of piRNAs offer an opportunity to better understand cancer-specific biology as well as develop novel prognostic markers for clinical application.
